MOLECULAR CLONING METHOD: DNA CLONING VECTORS
MOLECULAR CLONING METHOD:DNA
CLONING VECTORS:-
INTRODUCTION:-
- DNA cloning is a cornerstone of molecular biology. A clone can be defined as an identical copy.
- DNA cloning involved joining or ligating.
- DNA with a vector that enables the resulting contract to be introduced into a cell, replicatedand passed on to daughter cells, as that cell divides.
- DNA exists as a double helix of antiparallel polymer strand with the nucleotide units in each joined by 5’-3’ phosphodiesterbond.
- The ability to selectively break and join these bind is the basis of DNA cloning.
WHAT IS DNA CLONING :-
- DNA cloning allows a copy of any specific part of DNA ( or RNA ) sequences to be selected among many others and produced in an unlimited amount.
- The recombinant DNA is produced by cloning a foregin DNA isolated either from the genome or synthesized chemically or as a cDNA library using mRNA molecules.
- DNA cloning allows a copy of any specific part of DNA ( or RNA ) sequences to be selected among many others and produced in an unlimited amount.
- The recombinant DNA is produced by cloning a foregin DNA isolated either from the genome or synthesized chemically or as cDNA using mRNA molecules.
- However the cloning of this DNA can be done only when the ' another DNA molecules' is available that may replicate in the transformed host cell.This other DNA molecules uses for joining the foregin DNA is called vector.
- This technique is the first stage of most of the genetic engineering experiments.
Production of DNA libraries
- PCR
- DNA sequencing
- The DNA cloning is the massive amplification of DNA sequence .
- DNA cloning is a stable propogation of DNA sequence.
- A single DNA molecules can be amplified allowing it to be :-
- Studied – sequenced
- Manipulated –mutagenised
- Expressed – generation of protein
Restrictions Enzymes :-
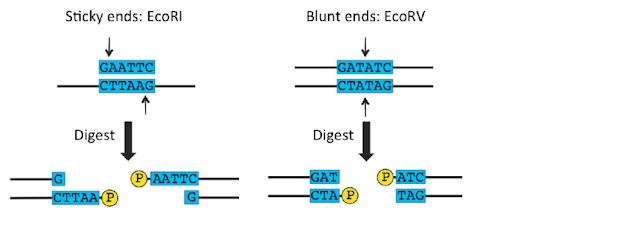
- The blunt end contains the same location from blunt end.
- E.g. EcoRV
- The sticky end also called the cohesive end .At the same site cleavage and forms the fragments it get sticky end.E.g. EcoRl and Taq l
- Restrictions endonuclease are of 4 types
- Type l
- Type ll
- Type lll
- Type ll s
- Type l R.E cut the DNA 1000 bp long away from the recognition site.It required ATP and mg ++ as a cofactor. It contains three subunit 1.
- Type ll restrictions endonuclease enzyme is commonly use enzyme. It does not require ATPbut mg ++ is use as a cofactor.Recognatiin site and cleavage site
- Type lll restrictions endonuclease cut the DNA at 24-26 base pair far away from recognition site.
- They require ATP and Mg ++ as cofactor.
- Type ll s it cleavage the modified DNA.
NOMENCLATURE OF
RESTRICTIONS ENDONUCLEASE:-
- First capital letter represent genus of the organism.
- Second two small letter represent species of organism.
- Next letter represent strains.
- Next Roman number represents order of discovery.E.g. EcoRl
CLONING VECTORS:-
INTRODUCTION :- A cloning vectors is a DNA molecules in which foregin DNA can be inserted and which is further capable of replicating within the host cell to product the multiple clones of recombinant DNA e.g. plasmids ,phage or viruses.
HISTORY:-
- Scientists working in Bayer’s lab recognised general cloning vectors with unique restriction site for cloning in foregin DNA.
- In 1977, they described this 1st vector designed for cloning purpose, pBR322-a plasmid.
- This vector was small, approximately 4 kb in size, and had two antibiotic resistance genes for selection.
USES:-
- Cloning vectors is used as a vehicle to artificially carry foregin genetic material into another cell, where it can replicate and expressed.
- Cloning vectors are DNA molecules that are used to transport cloned sequence between biological host and the test tubes.
FEATURES OF CLONING VECTORS:-
ORIGIN OF REPLICATION:-
- This marker is autonomous replication in vector.
- Ori is a specific sequence of nucleotide from where replication starts.
- When the foregin DNA is linked to the sequence along with vector replication, foreign DNA also starts replicating within host.
CLONING SITES:-
- Cloning site is a place where the vector DNA can be digested and desired DNA can be inserted by the same restrictions enzyme.
- It is a point analysis for genetic engineering.
- Recently recombinant plasmid contains a multiple Cloning site which has many restriction site.
SELECTABLE MARKER:-
- Selectable marker is a gene that contains resistance to a particular antibiotic agents that would normally kill the host cell.
- A cloning vectors contains a selectable marker which confer on the host cell an ability to survive and proliferate in a selective growth medium containing the particular antibiotic.
PLASMID:-
INTRODUCTION:-
Extra chromosomal DNA molecules,
- Self replicating
- Double stranded
- Short sequence of DNA
- Circular DNA molecules.
- CHARACTERISTICS:-
- Minimum amount of DNA
- Two suitable marker for identification
- Relaxed replication control
- Restrictions endonuclease enzyme
TYPE
3:-
- FERTILITY PLASMID:- can perform conjugation
RESISTANCE PLASMIDS: - contains gene that build a resistance against antibiotic.
COL PLASMID: - Contains gene that code for protein that can kill bacteria.
Plasmid PBR322:-
- The PBR322 is an artificial plasmid.
- Its DNA is derived from three different but naturally occurring plasmid.
- The size of PBR322 is 2.9 M Da.
- It is isolated from E.coli strain RPI.
- It contains gene that give resistance against two antibiotics namely ampicillin and tetracycline.
- The plasmid has restrictions site of over 20 RE.
NOMENCLATURE:-
- P – plasmid
- BR- Bolivar Rodriguez
- 322- It is number given to distinguish the plasmid.
ORIGIN OF PLASMID:-
- Gene ampicillin resistance = RSG 2124
- Gene for tetracycline resistance = psc 101
- Origin of replication = PMB
ADVANTAGES OF PBR322:-
- This is the most commonly used plasmid in gene cloning experiments.
- It is very smaller than other natural plasmid.
- In this plasmid 6 kb DNA can be inserted.
pUC18 vector :-
- pUC19 is
one of a series of plasmid cloning
vectors created by Joachim
Messing and co-workers.
- The designation "pUC" is
derived from the classical "p" prefix (denoting "plasmid") and the abbreviation for the University of California, where early work on the plasmid series had been conducted.
- It is a circular double stranded DNA and has 2686 base pairs.
- pUC19 is one of the most widely used
vector molecules as the recombinants, or the cells into which foreign DNA has been introduced,
can be easily distinguished from the non-recombinants based on color
differences of colonies on growth media.
- pUC18 is similar to pUC19, but the MCS region is reversed.
BACTERIOPHAGE VECTORS :-
- BACTERIOPHAGE is the viruses that infect the bacterial cells after injecting their genetic material and kill them.
- The viral DNA expressed and replicate inside the bacterial cells and produce the number of phage particles released after bursting the bacterial cells.
- This is called lytic cycle of bacteriophage.
- The two BACTERIOPHAGE e.g. .lambda phage and M13 have been modified.
M13 BACTERIOPHAGE:-
M13 is a filamentous bacteriophage composed of circular single-stranded DNA (ssDNA) which is 6407 nucleotides long encapsulated in approximately 2700 copies of the major coat protein P8, and capped with 5 copies of two different minor coat proteins (P9, P6, P3) on the ends.
The minor coat protein P3 attaches to the receptor at the tip of the F pilus of the host Escherichia coli. Infection with M13 lysogenic phage.
The infection causes turbid plaques in E. coli lawns, of intermediary opacity in comparison to regular lysis plaques.
However, a decrease in the rate of cell growth is seen in the infected cells. M13 plasmids are used for many recombinant DNA processes, and the virus has also used for phage display, nanostructures and nanotechnology applications.
Replication in E. coli:-
Below are steps involved with
replication of M13 in E. coli.
Viral (+) strand DNA enters cytoplasm
Complementary (-) strand is synthesized
by bacterial enzymes
DNA Gyrase, a type II topoisomerase,
acts on double-stranded DNA and catalyzes formation of negative super coils in
double-stranded DNA
Final product is parental replicative
form (RF) DNA
A phage protein, pII, nicks the (+)
strand in the RF
3'-hydroxyl acts as a primer in the
creation of new viral strand
pIIcirculizes displaced viral (+)
strand DNA
Pool of progeny double-stranded RF
molecules produced.
Negative strand of RF is
template of transcription
mRNAs are translated into the phage proteins.
LAMBDA
PHAGE:-
Commonly known as vector.
Lambda phage specially infects the E.coli.
DNA of lambda phage is specially 48.5 kb in length .
As it’s ends are the cos site which consists of 12 bp of cohesive end.
The cos end allow the DNA to be circularized in the host cell.
For cloning of large DNA fragments up to 20 bp non essential lambda DNA is removed and replaced and inserted in it.
Recombinant DNA is packed within viral particles these are allow to infect the bacterial cell.
COSMID :-
A cosmid is a type of hybrid plasmid that contains a Lambda phage cos sequence . Cosmids (cos sites + plasmid = cosmids) DNA sequences are originally from the lambda phage.
They are often used as a cloning vector in genetic engineering. Cosmids can be used to build genomic libraries.
They can replicate as plasmids if they have a suitable origin of replication (ori): for example SV40 ori in mammalian cells, ColE1 ori for double-stranded DNA replication, or f1 ori for single-stranded DNA replication in prokaryotes.
COSMID
FEATURES AND USES :-
Cosmids are predominantly plasmids with a bacterial oriV, an antibiotic selection marker and a cloning site, but they carry one, or more recently two, cos sites derived from bacteriophage lambda.
Depending on the particular aim of the experiment, broad host range cosmids, shuttle cosmids or 'mammalian' cosmids (linked to SV40 oriV and mammalian selection markers) are available.
The loading capacity of cosmids varies depending on the size of the vector itself but usually lies around 40–45 kb.
PHAGEMID VECTORS:-
Sometime generation of a single stranded DNA becomes important for DNA containing bacteriophage M13 .
A phagemid or phasmid is a DNA-based cloning vector, which has both bacteriophageand plasmid properties.
These vectors carry, in addition to the origin of plasmid replication, an origin of replication derived from bacteriophage.
Unlike commonly used plasmids, phagemid vectors differ by having the ability to be packed into the capsid of a bacteriophage due to their having a genetic sequence that signals for packaging .
Phagemids are used in a variety of biotechnology applications; for example, they can be used in a molecular biology technique called "Phage Display.
A phagemid is a plasmid that contains an f1 origin of replication from an f1 phage.
It can be used as a type of cloning vector in combination with filamentous phage M13.
A phagemid can be replicated as a plasmid, and also be packaged as single stranded DNA in viral particles
. Phagemids contain an origin of replication (ori) for double stranded replication, as well as an f1 ori to enable single stranded replication and packaging into phage particles.
- Many commonly used plasmids contain an f1 ori and are thus phagemids.




















No comments:
Post a Comment